Absolute individual weighted residuals vs population predictions or independent variable for Xpose 4
Source:R/absval.iwres.vs.pred.R
absval.iwres.vs.pred.RdThis is a plot of absolute individual weighted residuals (|IWRES|) vs
individual predictions (PRED) or independent variable (IDV), specific
functions in Xpose 4. These functions are wrappers encapsulating arguments
to the xpose.plot.default function. Most of the options take their
default values from xpose.data object but may be overridden by supplying
them as arguments.
Usage
absval.iwres.vs.pred(
object,
ylb = "|IWRES|",
smooth = TRUE,
idsdir = "up",
type = "p",
...
)Arguments
- object
An xpose.data object.
- ylb
A string giving the label for the y-axis.
NULLif none.- smooth
Logical value indicating whether an x-y smooth should be superimposed. The default is TRUE.
- idsdir
Direction for displaying point labels. The default is "up", since we are displaying absolute values.
- type
Type of plot. The default is points only ("p"), but lines ("l") and both ("b") are also available.
- ...
Other arguments passed to
link{xpose.plot.default}.
Details
A wide array of extra options controlling xyplots are available. See
xpose.plot.default for details.
See also
xpose.plot.default,
xpose.panel.default, xyplot,
xpose.prefs-class, xpose.data-class
Other specific functions:
absval.cwres.vs.cov.bw(),
absval.cwres.vs.pred(),
absval.cwres.vs.pred.by.cov(),
absval.iwres.cwres.vs.ipred.pred(),
absval.iwres.vs.cov.bw(),
absval.iwres.vs.idv(),
absval.iwres.vs.ipred(),
absval.iwres.vs.ipred.by.cov(),
absval.wres.vs.cov.bw(),
absval.wres.vs.idv(),
absval.wres.vs.pred(),
absval.wres.vs.pred.by.cov(),
absval_delta_vs_cov_model_comp,
addit.gof(),
autocorr.cwres(),
autocorr.iwres(),
autocorr.wres(),
basic.gof(),
basic.model.comp(),
cat.dv.vs.idv.sb(),
cat.pc(),
cov.splom(),
cwres.dist.hist(),
cwres.dist.qq(),
cwres.vs.cov(),
cwres.vs.idv(),
cwres.vs.idv.bw(),
cwres.vs.pred(),
cwres.vs.pred.bw(),
cwres.wres.vs.idv(),
cwres.wres.vs.pred(),
dOFV.vs.cov(),
dOFV.vs.id(),
dOFV1.vs.dOFV2(),
data.checkout(),
dv.preds.vs.idv(),
dv.vs.idv(),
dv.vs.ipred(),
dv.vs.ipred.by.cov(),
dv.vs.ipred.by.idv(),
dv.vs.pred(),
dv.vs.pred.by.cov(),
dv.vs.pred.by.idv(),
dv.vs.pred.ipred(),
gof(),
ind.plots(),
ind.plots.cwres.hist(),
ind.plots.cwres.qq(),
ipred.vs.idv(),
iwres.dist.hist(),
iwres.dist.qq(),
iwres.vs.idv(),
kaplan.plot(),
par_cov_hist,
par_cov_qq,
parm.vs.cov(),
parm.vs.parm(),
pred.vs.idv(),
ranpar.vs.cov(),
runsum(),
wres.dist.hist(),
wres.dist.qq(),
wres.vs.idv(),
wres.vs.idv.bw(),
wres.vs.pred(),
wres.vs.pred.bw(),
xpose.VPC(),
xpose.VPC.both(),
xpose.VPC.categorical(),
xpose4-package
Examples
if (FALSE) { # \dontrun{
## We expect to find the required NONMEM run and table files for run
## 5 in the current working directory
xpdb5 <- xpose.data(5)
} # }
## Here we load the example xpose database
data(simpraz.xpdb)
xpdb <- simpraz.xpdb
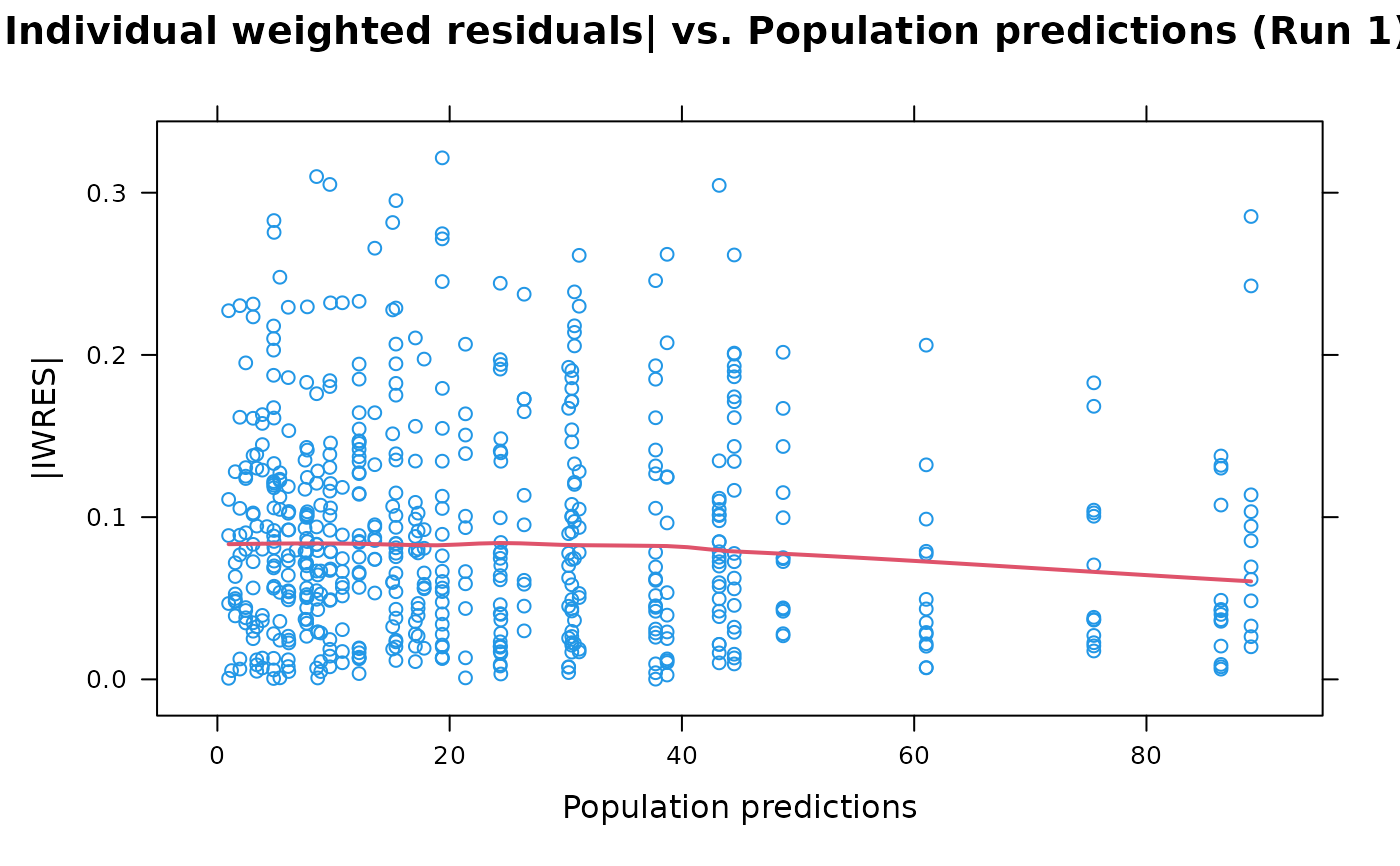
## A vanilla plot
absval.iwres.vs.pred(xpdb)
 ## A conditioning plot
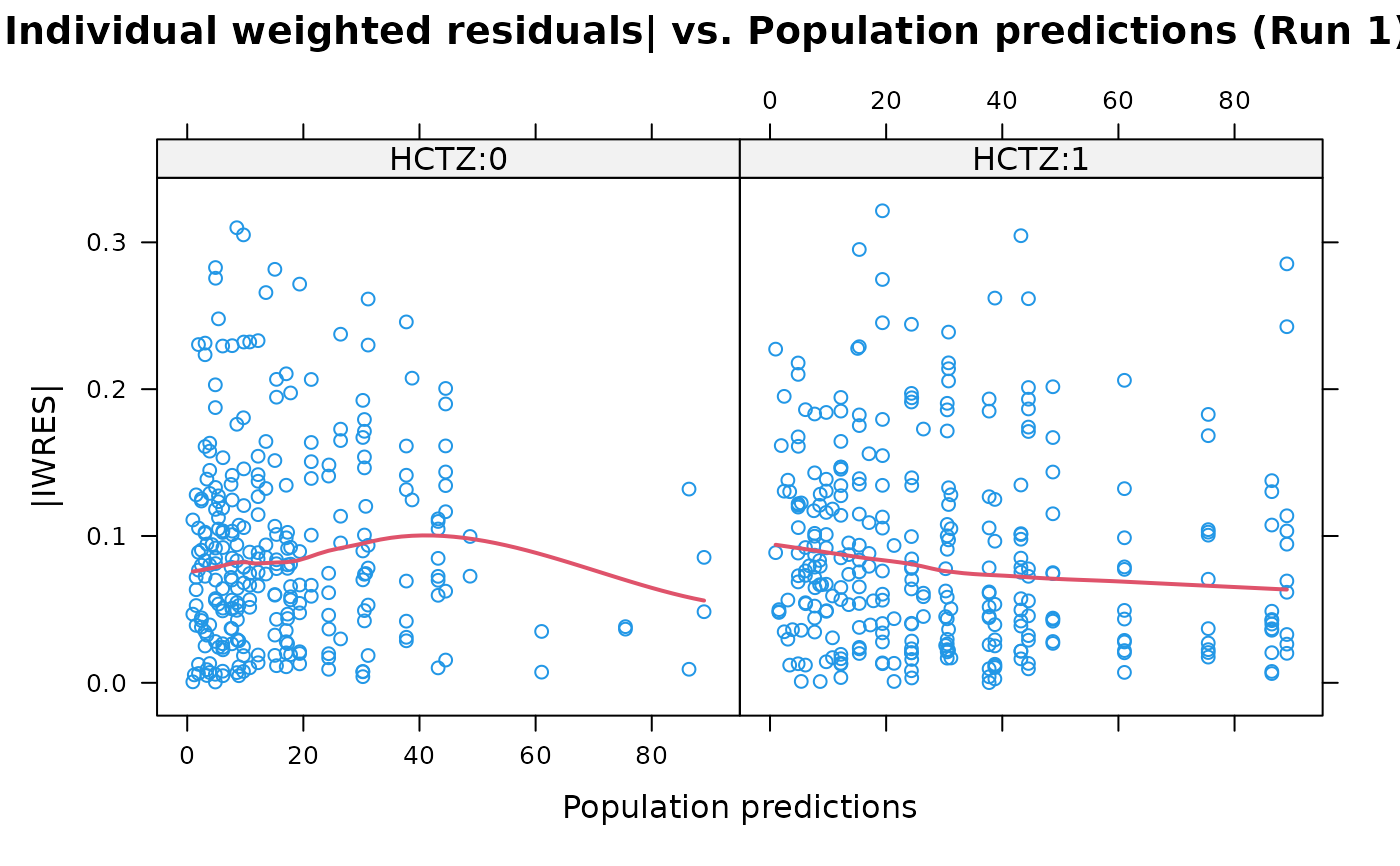
absval.iwres.vs.pred(xpdb, by="HCTZ")
## A conditioning plot
absval.iwres.vs.pred(xpdb, by="HCTZ")
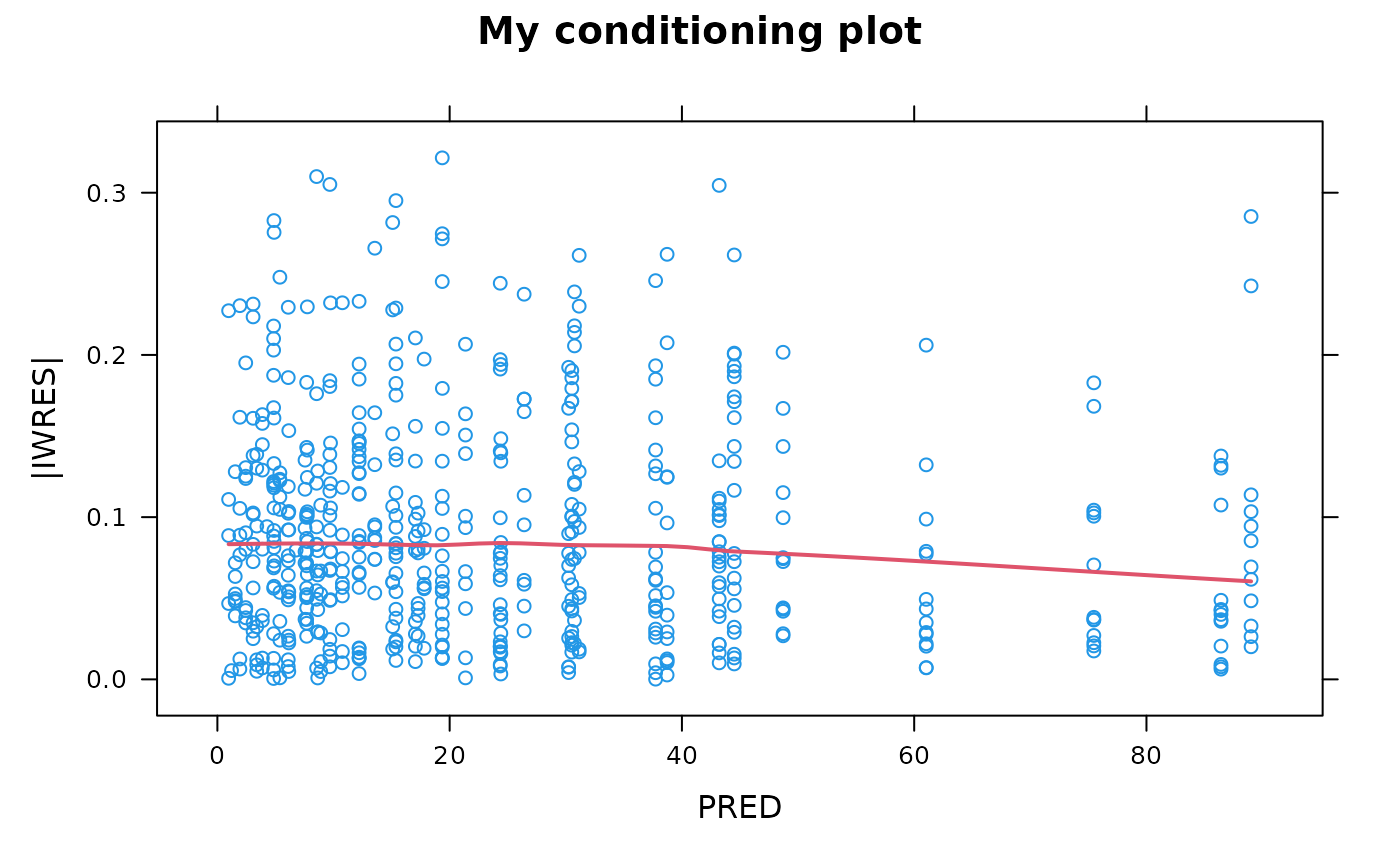
 ## Custom heading and axis labels
absval.iwres.vs.pred(xpdb, main="My conditioning plot", ylb="|IWRES|", xlb="PRED")
## Custom heading and axis labels
absval.iwres.vs.pred(xpdb, main="My conditioning plot", ylb="|IWRES|", xlb="PRED")
 ## Custom colours and symbols, no IDs
absval.iwres.vs.pred(xpdb, cex=0.6, pch=3, col=1, ids=FALSE)
## Custom colours and symbols, no IDs
absval.iwres.vs.pred(xpdb, cex=0.6, pch=3, col=1, ids=FALSE)
